Success is in the detail
“The devil is in the detail” is a saying often used to point to unexpected events in a process. In other words, one can say “success is in the detail”.
Welcome to INSIGNIA-EU, an exciting new pan-european citizen scientist study with beekeepers. The Mission statement of the is INSIGNIA -EU study is: “INSIGNIA-EU will extend the INSIGNIA pilot project (2019-2021) to encompass pan-European environmental pollution bio-monitoring using honey bee colonies. The main pillars are: 1) Robust data, generated by beekeeper citizen scientists via a clear bio-monitoring guideline (protocol), new simple and easy to handle sampling techniques and sample preservation methods, quality controlled sample analyses to low levels and professional data handling; and 2) Modelling of land use – related to environmental (established and emergent) pollutants, pesticide exposure risk for honey bees and pollen diversity.”. Continue reading “INSIGNIA-EU, success is in details”

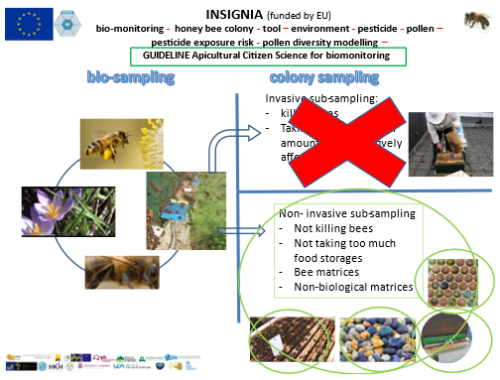
 The INSIGNIA study has been running now for two years and we are entering the final stage. In these two years, we have gone through all stages of organizing an apiculturist citizen science study. We developed the passive sampler APIStrip, the efficient matrix to capture pesticides, and all the pitfalls that came on the way like the “not-to-be -misinterpretable” label, best practice for storage shipping, sampling schemes etc. Now it is time to bring all of this information together in the “Guideline for apiculturist citizen science for applying honeybee colonies for bio-monitoring of the environment – subject pesticides”. Given the Covid-19 restrictions, physical meetings to discuss all of the stages, best wording, most logical set-up, best flow-charts, and whatever it takes to write the best guidelines, would be great but are sadly impossible. Therefore we have started a scheme of virtual Teams-meetings with a strict agenda to go through all of these aspects. We started in December 2020 and will be finished in March 2021. There will be as many Teams-meetings as it takes to get the job done in time.
The INSIGNIA study has been running now for two years and we are entering the final stage. In these two years, we have gone through all stages of organizing an apiculturist citizen science study. We developed the passive sampler APIStrip, the efficient matrix to capture pesticides, and all the pitfalls that came on the way like the “not-to-be -misinterpretable” label, best practice for storage shipping, sampling schemes etc. Now it is time to bring all of this information together in the “Guideline for apiculturist citizen science for applying honeybee colonies for bio-monitoring of the environment – subject pesticides”. Given the Covid-19 restrictions, physical meetings to discuss all of the stages, best wording, most logical set-up, best flow-charts, and whatever it takes to write the best guidelines, would be great but are sadly impossible. Therefore we have started a scheme of virtual Teams-meetings with a strict agenda to go through all of these aspects. We started in December 2020 and will be finished in March 2021. There will be as many Teams-meetings as it takes to get the job done in time.